CSD Cross Miner Terminology
CSD-Cross Miner can be thought of as a pharmacophore based query tool.
However, it is much more powerful than traditional pharmacophore query tools
as it allows you to query not only databases of ligands, but also proteins and
protein-ligand interactions. CSD-CrossMiner includes a preconfigured database
of biologically relevant subsets of the Cambridge Structural Database (CSD) and
the Protein Data Bank (PDB). The pharmacophore used in the query is interactive,
allowing you to easily edit it and in a number of ways through a simple user
interface. This delivers an overall interactive search experience with application
areas in interaction searching, scaffold hopping or the identification of novel
fragments for specific protein environments.
The subset of the CSD included with CSD-CrossMiner consists of structures which
are not organometallic, have an R-factor of at maximum 10%, have 3D
coordinates, have no disorder, and are not polymeric (about 285 437 structures
total). The included PDB database is a subset of all co-crystalized ligands, with
the binding site defined as all molecules with an atom within a 6Å radius around
the ligand (205 194 binding sites)
CSD-Cross Miner Terminology
CSD-CrossMiner uses several terms, some common to the field of drug
discovery, and some not. For reference, these terms are defined as below:
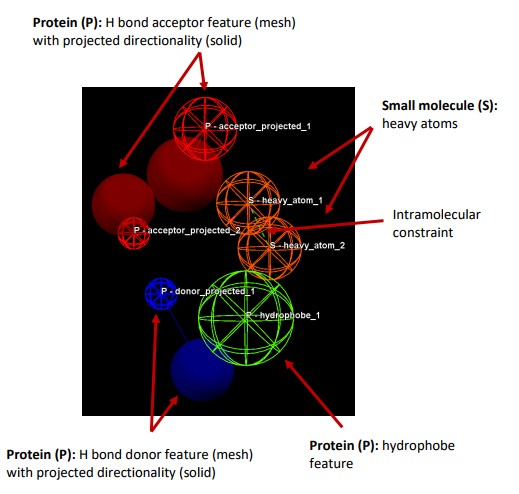
Features: can be defined as an ensemble of steric and electronic features
that characterise a protein and/or a small molecule. In CSD-CrossMiner a
feature is defined as point(s), centroid or vector which represent a SMARTS
query and, in the case of a vector, this includes geometric rules.
Pharmacophore point: is a feature that has been selected to be part of a
pharmacophore because its presence is necessary to ensure the optimal
supramolecular interactions with a specific CSD-Cross Miner Terminology
CSD-CrossMiner uses several terms, some common to the field of drug
discovery, and some not. For reference, these terms are defined as below:
Features: can be defined as an ensemble of steric and electronic features
that characterise a protein and/or a small molecule. In CSD-CrossMiner a
feature is defined as point(s), centroid or vector which represent a SMARTS
query and, in the case of a vector, this includes geometric rules.
Pharmacophore point: is a feature that has been selected to be part of a
pharmacophore because its presence is necessary to ensure the optimal
supramolecular interactions with a specific biological target and to trigger or
block its biological response.
Structure database: is a database containing the 3D coordinates of small
molecule structures and/or protein-ligand binding sites. This database is
used to create a feature database.
Feature database: is a database containing the structures from the structure
database, indexed with a set of feature definitions provided by CSDCrossMiner
and any additional features defined by the user. This is the
database that CSD-CrossMiner uses to perform the actual 3D search against
a pharmacophore query.
Exit vector: is a two-point feature that represents a single, non-ring bond
between two heavy atoms features, and it will be represented as two mesh
spheres. In the case of CSD-CrossMiner, directionality in an exit vector does
not matter. target and to trigger or
block its biological response.
Structure database: is a database containing the 3D coordinates of small
molecule structures and/or protein-ligand binding sites. This database is
used to create a feature database.
Feature database: is a database containing the structures from the structure
database, indexed with a set of feature definitions provided by CSDCrossMiner
and any additional features defined by the user. This is the
database that CSD-CrossMiner uses to perform the actual 3D search against
a pharmacophore query.
Exit vector: is a two-point feature that represents a single, non-ring bond
between two heavy atoms features, and it will be represented as two mesh
spheres. In the case of CSD-CrossMiner, directionality in an exit vector does
not matter.

Leave a comment